Determine sample sizes for a set of algorithms on a single problem instance
Source:R/calc_nreps.R
calc_nreps.RdIteratively calculates the required sample sizes for K algorithms on a given problem instance, so that the standard errors of the estimates of the pairwise differences in performance is controlled at a predefined level.
calc_nreps(
instance,
algorithms,
se.max,
dif = "simple",
comparisons = "all.vs.all",
method = "param",
nstart = 20,
nmax = 1000,
seed = NULL,
boot.R = 499,
ncpus = 1,
force.balanced = FALSE,
load.folder = NA,
save.folder = NA
)Arguments
- instance
a list object containing the definitions of the problem instance. See Section
Instancefor details.- algorithms
a list object containing the definitions of all algorithms. See Section
Algorithmsfor details.- se.max
desired upper limit for the standard error of the estimated difference between pairs of algorithms. See Section
Pairwise Differencesfor details.- dif
type of difference to be used. Accepts "perc" (for percent differences) or "simple" (for simple differences)
- comparisons
type of comparisons being performed. Accepts "all.vs.first" (in which cases the first object in
algorithmsis considered to be the reference algorithm) or "all.vs.all" (if there is no reference and all pairwise comparisons are desired).- method
method to use for estimating the standard errors. Accepts "param" (for parametric) or "boot" (for bootstrap)
- nstart
initial number of algorithm runs for each algorithm. See Section
Initial Number of Observationsfor details.- nmax
maximum total allowed number of runs to execute. Loaded results (see
load.folderbelow) do not count towards this total.- seed
seed for the random number generator
- boot.R
number of bootstrap resamples to use (if
method == "boot")- ncpus
number of cores to use
- force.balanced
logical flag to force the use of balanced sampling for the algorithms on each instance
- load.folder
name of folder to load results from. Use either "" or "./" for the current working directory. Accepts relative paths. Use
NAfor not saving.calc_nreps()will look for a .RDS file with the same name- save.folder
name of folder to save the results. Use either "" or "./" for the current working directory. Accepts relative paths. Use
NAfor not saving.
Value
a list object containing the following items:
instance- alias for the problem instance consideredXk- list of observed performance values for allalgorithmsNk- vector of sample sizes generated for each algorithmDiffk- data frame with point estimates, standard errors and other information for all algorithm pairs of interestseed- seed used for the PRNGdif- type of difference usedmethod- method used ("param" / "boot")comparisons- type of pairings ("all.vs.all" / "all.vs.first")
Instance
Parameter instance must be a named list containing all relevant parameters
that define the problem instance. This list must contain at least the field
instance$FUN, with the name of the function implementing the problem
instance, that is, a routine that calculates y = f(x). If the instance
requires additional parameters, these must also be provided as named fields.
Algorithms
Object algorithms is a list in which each component is a named
list containing all relevant parameters that define an algorithm to be
applied for solving the problem instance. In what follows algorithm[[k]]
refers to any algorithm specified in the algorithms list.
algorithm[[k]] must contain an algorithm[[k]]$FUN field, which is a
character object with the name of the function that calls the algorithm; as
well as any other elements/parameters that algorithm[[k]]$FUN requires
(e.g., stop criteria, operator names and parameters, etc.).
The function defined by the routine algorithm[[k]]$FUN must have the
following structure: supposing that the list in algorithm[[k]] has
fields algorithm[[k]]$FUN = "myalgo", algorithm[[k]]$par1 = "a" and
algorithm$par2 = 5, then:
myalgo <- function(par1, par2, instance, ...){
# do stuff
# ...
return(results)
}
That is, it must be able to run if called as:
# remove '$FUN' and '$alias' fields from list of arguments
# and include the problem definition as field 'instance'
myargs <- algorithm[names(algorithm) != "FUN"]
myargs <- myargs[names(myargs) != "alias"]
myargs$instance <- instance
# call function
do.call(algorithm$FUN,
args = myargs)
The algorithm$FUN routine must return a list containing (at
least) the performance value of the final solution obtained, in a field named
value (e.g., result$value) after a given run.
Initial Number of Observations
In the general case the initial number of observations per algorithm
(nstart) should be relatively high. For the parametric case
we recommend between 10 and 20 if outliers are not expected, or between 30
and 50 if that assumption cannot be made. For the bootstrap approach we
recommend using at least 20. However, if some distributional assumptions can
be made - particularly low skewness of the population of algorithm results on
the test instances), then nstart can in principle be as small as 5 (if the
output of the algorithms were known to be normal, it could be 1).
In general, higher sample sizes are the price to pay for abandoning
distributional assumptions. Use lower values of nstart with caution.
Pairwise Differences
Parameter dif informs the type of difference in performance to be used
for the estimation (\(\mu_a\) and \(\mu_b\) represent the mean
performance of any two algorithms on the test instance, and \(mu\)
represents the grand mean of all algorithms given in algorithms):
If
dif == "perc"andcomparisons == "all.vs.first", the estimated quantity is \(\phi_{1b} = (\mu_1 - \mu_b) / \mu_1 = 1 - (\mu_b / \mu_1)\).If
dif == "perc"andcomparisons == "all.vs.all", the estimated quantity is \(\phi_{ab} = (\mu_a - \mu_b) / \mu\).If
dif == "simple"it estimates \(\mu_a - \mu_b\).
References
F. Campelo, F. Takahashi: Sample size estimation for power and accuracy in the experimental comparison of algorithms. Journal of Heuristics 25(2):305-338, 2019.
P. Mathews. Sample size calculations: Practical methods for engineers and scientists. Mathews Malnar and Bailey, 2010.
A.C. Davison, D.V. Hinkley: Bootstrap methods and their application. Cambridge University Press (1997)
E.C. Fieller: Some problems in interval estimation. Journal of the Royal Statistical Society. Series B (Methodological) 16(2), 175–185 (1954)
V. Franz: Ratios: A short guide to confidence limits and proper use (2007). https://arxiv.org/pdf/0710.2024v1.pdf
D.C. Montgomery, C.G. Runger: Applied Statistics and Probability for Engineers, 6th ed. Wiley (2013)
Examples
# Example using dummy algorithms and instances. See ?dummyalgo for details.
# We generate dummy algorithms with true means 15, 10, 30, 15, 20; and true
# standard deviations 2, 4, 6, 8, 10.
algorithms <- mapply(FUN = function(i, m, s){
list(FUN = "dummyalgo",
alias = paste0("algo", i),
distribution.fun = "rnorm",
distribution.pars = list(mean = m, sd = s))},
i = c(alg1 = 1, alg2 = 2, alg3 = 3, alg4 = 4, alg5 = 5),
m = c(15, 10, 30, 15, 20),
s = c(2, 4, 6, 8, 10),
SIMPLIFY = FALSE)
# Make a dummy instance with a centered (zero-mean) exponential distribution:
instance = list(FUN = "dummyinstance", distr = "rexp", rate = 5, bias = -1/5)
# Explicitate all other parameters (just this one time:
# most have reasonable default values)
myreps <- calc_nreps(instance = instance,
algorithms = algorithms,
se.max = 0.05, # desired (max) standard error
dif = "perc", # type of difference
comparisons = "all.vs.all", # differences to consider
method = "param", # method ("param", "boot")
nstart = 15, # initial number of samples
nmax = 1000, # maximum allowed sample size
seed = 1234, # seed for PRNG
boot.R = 499, # number of bootstrap resamples (unused)
ncpus = 1, # number of cores to use
force.balanced = FALSE, # force balanced sampling?
load.folder = NA, # file to load results from
save.folder = NA) # folder to save results
#>
#> Sampling algorithms on instance dummyinstance : .....................................
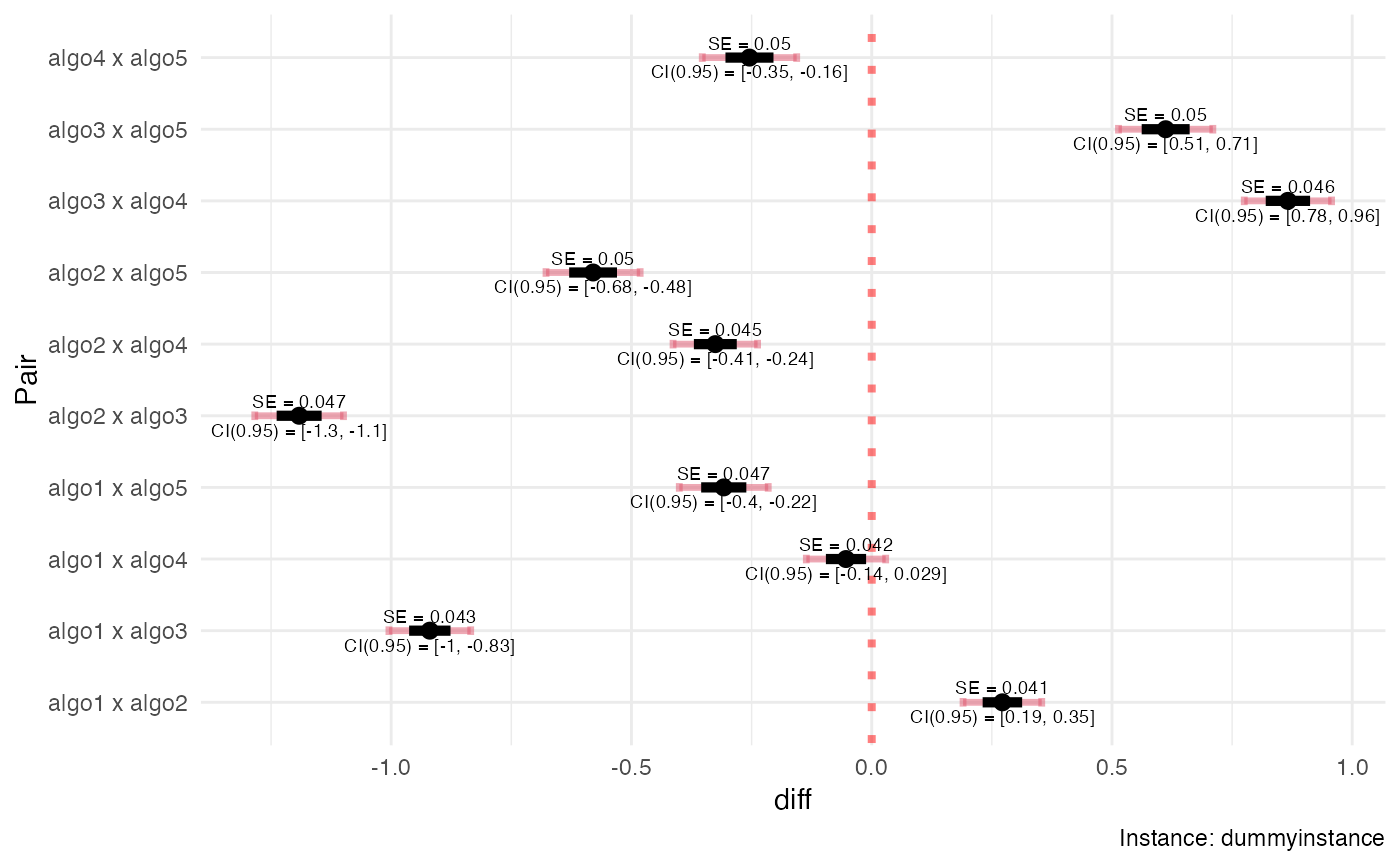
summary(myreps)
#> #====================================
#> Instance: dummyinstance
#> Number of algorithms: 5
#> algo1: 15 runs
#> algo2: 61 runs
#> algo3: 138 runs
#> algo4: 165 runs
#> algo5: 237 runs
#> --------------------
#> Total runs: 616
#> Comparisons: all.vs.all
#> #====================================
#>
#> Alg1 Alg2 N1 N2 Phi SE r ropt
#> 1 1 2 15 61 0.2720 0.0411 0.2460 0.435
#> 2 1 3 15 138 -0.9200 0.0431 0.1090 0.288
#> 3 1 4 15 165 -0.0535 0.0418 0.0909 0.254
#> 4 1 5 15 237 -0.3080 0.0470 0.0633 0.177
#> 5 2 3 61 138 -1.1900 0.0468 0.4420 0.662
#> 6 2 4 61 165 -0.3250 0.0446 0.3700 0.585
#> 7 2 5 61 237 -0.5800 0.0498 0.2570 0.408
#> 8 3 4 138 165 0.8660 0.0462 0.8360 0.883
#> 9 3 5 138 237 0.6120 0.0499 0.5820 0.617
#> 10 4 5 165 237 -0.2540 0.0500 0.6960 0.698
#>
#> #====================================
plot(myreps)